Metabolome Informatics Research Team, RIKEN Center for Sustainable Resource Science (CSRS)
MetaboBank, a metabolomics repository
— Creating the means for archiving and reusing metabolite data —
MetaboBank, a metabolomics repository for comprehensively analyzing metabolites, was established in 2021. The mission of the facility is to collect metabolomics data much like physical specimens are collected, storing this data permanently as a resource that others can access and use for re-analysis and other purposes. Team Leader Masanori Arita, who designed and built MetaboBank, spoke to us about the importance of preserving metabolite data and future plans for the MetaboBank.

Masanori Arita
Team Leader
Metabolome Informatics Research Team, RIKEN CSRS
Advances in mass spectrometry technology driving metabolomics research
Many different metabolites are produced within the body as a result of life activities. These metabolites are referred to collectively as “metabolomes,” and the research field and techniques for comprehensively analyzing metabolomes is known as “metabolomics.”
Metabolomics is the third “-omics” field following genomics and proteomics. Metabolomics research began to take off around 2000, driven by advances in mass spectrometry technology. The body produces myriad metabolites, including sugars, amino acids, organic acids, lipids, and vitamins. Metabolomics involves the simultaneous, comprehensive detection and measurement of the mass of compounds with a molecular weight of approximately 1500 or less. Compared to genome and proteome analysis, metabolome analysis poses various technical hurdles but advances in mass spectrometry technology have enabled such hurdles to be surmounted.
However, state-of-the-art mass spectrometers are very expensive and require specialized skills. Samples may also be difficult to obtain in the case of rare organisms. These circumstances have prompted the establishment of public repositories in Europe and the USA to centrally store metabolomics data and make it available to anyone. Japan was a little slow to follow such leads, but in 2021 we launched MetaboBank as the first public metabolomics repository in Asia.
Ensuring the permanent storage and reuse of data
MetaboBank aims to permanently store metabolomics data and facilitate use of this data by others. To this end, MetaboBank is managed under the jurisdiction of the DNA Data Bank of Japan (DDBJ), a globally recognized repository of nucleotide sequences with a history of almost 40 years. In addition to my position in RIKEN, I also serve as director of the DDBJ Center at the National Institute of Genetics and have been involved in the design and construction of MetaboBank. Data management and other everyday operations are carried out by curators.
Enabling reuse of the MetaboBank’s data requires the inclusion of metadata that explains what the data is. Metabolomics involves measuring diverse types of compounds, and samples such as fermented foods may contain a mixture of plant and bacterial compounds. This means that various methods are required to extract and measure samples, which in turn requires metadata regarding methods used to prepare and measure specific types of samples.
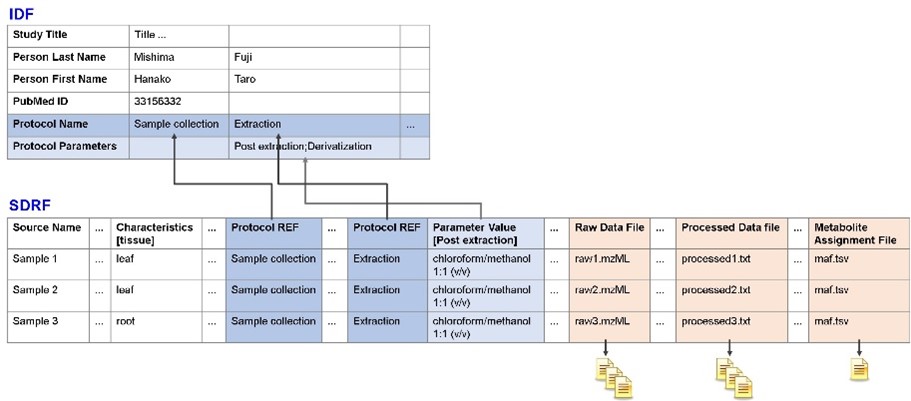
Fig.1: Relationship between IDF, SDRF, raw and analyzed data files, and MAF in MAGE-TAB
MetaboBank’s metadata is in the MAGE-TAB format that is also used in genomics and proteomics.
Increasing the number of items to be recorded in the metadata increases the amount of work required and makes registering the data more difficult; cutting the number of metadata items risks making the metabolome data unusable. Getting this balance right is something that we have struggled with. Once we had drawn up a standardized registration method, we were finally able to start operating MetaboBank. The MetaboBank website provides detailed registration procedures, and we hope that interested parties will register their data in line with these procedures.
(MetaboBank registration: https://www.ddbj.nig.ac.jp/metabobank/submission-e.html)
Since MetaboBank was established recently, the number of metabolomes registered is still small, but our efforts to publicize the facility are paying off, and the amount of data is gradually increasing. We are wooing not only research institutions but also companies in the food industry and other sectors to register any metabolomes they might have. Riken has one of Japan’s top metabolomics research infrastructures, and data produced by the RIKEN Center for Sustainable Resource Science is registered in MetaboBank. We hope that anyone interested will download and use it.
Enabling data to be used like specimens in decades to come
Metabolomics is being applied not only to basic research but also increasingly to medicine, agriculture, food, environment, and other fields. Demand for metabolome data is also steadily increasing. Preserving such data properly is vital if, for example, 50 years from now, we need to know the state of the environment or quality of food in 2024. Future technologies may also be able to interpret current data in a way that leads to new discoveries. Moreover, if we fail to preserve data on endangered species now, we will have no way of knowing what metabolites they had if they became extinct.
Science has always been passed on to future generations by preserving specimens to record nature. Research data is an intellectual treasure and preserving it is vital to the future of science. In these competitive times, the priority is on publishing research papers, but most such papers do not on their own leave reproducible data. The data generated by your research can also be lost. I fervently hope that researchers pay serious thought to what will remain in 50 years’ time, why they are doing science, and go about their research with a firm eye on the future.
Another major issue is the cost of managing and maintaining the repository. Currently, both MetaboBank and DDBJ are under the administration of the National Institute of Genetics, but the volume of data is growing year by year; there is a limit to how much we can handle. These repositories of knowledge are a national asset, and I think we need to discuss how we can best maintain them.
(Article by: Chisato Hata/Photo by: Tadashi Aizawa/Production assistance: Sci-Tech Communications)
Links
- MetaboBank, a metabolomics public repository
Book
- Gakujutsu Shuppan no Kita Michi (“How Academic Publishing Has Evolved”) by Masanori Arita, pub. Iwanami Shoten, 2021 (Japanese only)







