Integrated Bioresource Information Division, RIKEN BioResource Research Center (BRC)
Taking the helm in bioresearch with the world’s largest store of bioresources and associated information
Bioresearch requires biological resources, also called bioresources. The RIKEN BioResource Research Center (RIKEN BRC) manages and distributes the world’s largest store of bioresources. A key to getting the most out of bioresources is maintaining information associated with them. I am researching how to maximize the synergy between bioresources and associated information.

Hiroshi Masuya
Director
Integrated Bioresource Information Division, RIKEN BRC
What is the RIKEN BRC?
The RIKEN BRC handles five types of bioresources: mice (approximately 9500 strains), plants (approximately 840,000 cell lines/clones), cells (approximately 17,000 lines), genes (approximately 3.8 million clones), and microorganisms (approximately 30,000 strains). No other institution around the globe handles bioresources at every single stage of life from organisms to cells and genes. Thus, the RIKEN BRC possesses the world’s largest store of bioresources.
As seen in how humans all differ in their constitution, organisms are diverse. However, with bioresources used in research, this diversity would preclude reproducibility in research, making it difficult to establish the scientific validity of results. Resolving this issue requires standard bioresources. One major mission of the BioResource Center is carefully controlling the quality of bioresources to prevent mutations.
Information is essential for eliciting the full potential of bioresources
I and the other members of the Integrated Bioresource Information Division specialize in biology and information science, and every day we research bioinformatics and ontology as experts. To coordinate and integrate the data put out by the bioresource centers around the world, we maintain and publish information with technology stipulated by international standards.
One method for doing so is the Resource Description Framework (RDF). The RDF is an international standard information description format that was devised to make the World Wide Web into a massive database of knowledge. For example, sentences describing bioresources are required to contain a subject, a predicate, and an object. A description of the subject and its object are followed by detailed information. The phrases used in these sentences are also carefully prescribed with consideration to ontology. These rules were established to enable computers to automatically associate information scattered across the Web and constantly update the database.
Reaping the benefits of these measures, the RIKEN BRC collaborates with databases around the world. For example, data from the Swiss Institute of Bioinformatics (SIB) on the homology of genes across species is integrated with the RIKEN BRC. Data provided by the RIKEN BRC, in turn, contributes to developing bioresource databases worldwide.
At the RIKEN BRC, we put effort into associating bioresources with disease information, and we link phenotype-related information to bioresources as well (Fig. 1). As in the case of, “If a certain phenotype is observed in mice, it is this disease”, phenotypes are often linked with diseases.
We also get creative with the user interface—that is, how to present this information. Using the search window at the upper-left of the RIKEN BRC homepage, users can search for information on all bioresources handled by the RIKEN BRC. Search results are displayed in an easy-to-read format by resource name, disease name, and gene. The site also permits searches by disease name. With comments such as, “Now that I understand how bioresources are associated with diseases, the scope of my research has expanded” and, “Being able to search across the five types of bioresources made it efficient”, users have voiced their delight.
Going forward, we also hope to link information such as the image database and brain database handled by the RIKEN Open Life Science Platform with the RDF. We are also considering use of AI to automatically link related data on the Web. We envision the creation of a “data ecosystem” through this sort of organic linking of information.

Fig. 1: Information provided by RIKEN BRC
Searching for “SHH” also allows the user to look up information on phenotypes such as deformed toes involving SHH.
Depositing and transferring bioresources and information
The RIKEN BRC does more than provide bioresources to researchers. Researchers can also deposit with and transfer to us the bioresources and information that they create.
When individual researchers manage their own bioresources and transfer them among each other, errors tend to occur, such as bioresources suddenly disappearing and becoming unavailable or being switched. When you deposit and transfer your bioresources with the RIKEN BRC, we control their quality and send out information in a manner that links it with the world. We hope you will deposit and transfer your bioresources with the RIKEN BRC so we can link your research with researchers around the globe.
We are also happy to receive information gained from studies that use bioresources. In some fields, the names of bioresources described in papers are sometimes unclear; and since we are not involved with the papers in question, we sometimes cannot link those bioresources with relevant information.
Our desire to propose bioresources based on information
Our work until now has focused on maintaining information associated with bioresources. As a new direction for our work, we are considering researching the production of bioresources from information (Fig. 2).
The RIKEN BRC not only provides bioresources but is also an information center that provides information. In addition, we research information gathering methods to efficiently link bioresources with massive data such as literature databases and DNA sequencing libraries. Gathering and analyzing large quantities of data on a daily basis yields findings such as, “There’s research momentum in this field” and, “This bioresource should be usable in other fields as well”. We hope to use these findings as a basis for producing new bioresources that we can then submit to the bioresearch world and thereby help to invigorate research. We will take on the challenge of actively developing bioresources.
Visit the RIKEN BRC homepage and try entering a keyword. What you discover may surprise you.
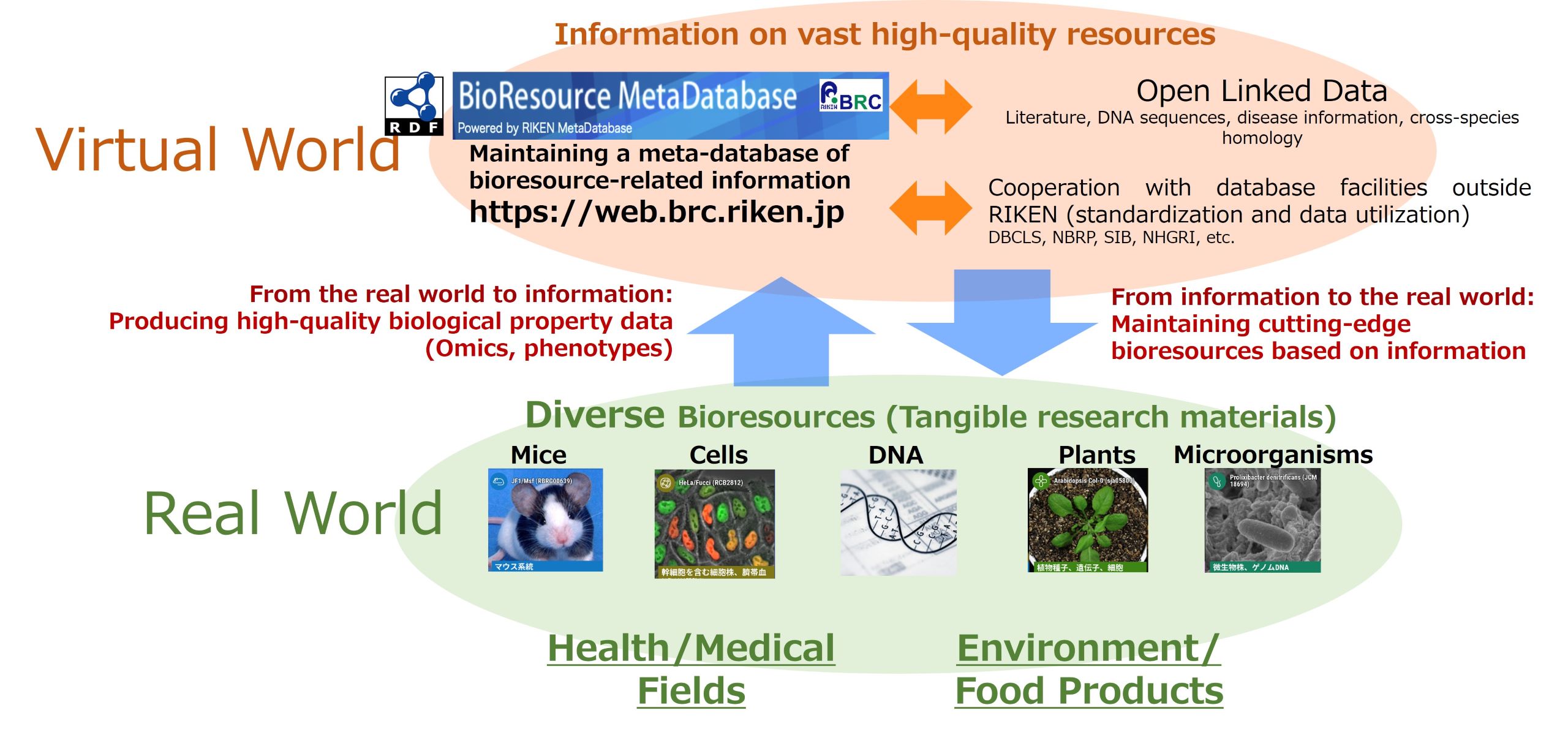
Fig. 2: Interactivity between information and bioresources

The members of the Integrated Bioresource Information Division, who leverage the RDF to send out information in collaboration with international databases (From left to right: Researcher Takada, Researcher Kushida, Division Director Masuya, Technical Staff member Usuda).
Links
-
Seach for bioresource on RIKEN BioResource Research Center (BRC)
- BRC Integrated Bioresource Information Division







